Running K2Taxonomer on Bulk Expression Data
Eric R. Reed
March 25, 2025
Source:vignettes/02_K2Taxonomer_bulk.Rmd
02_K2Taxonomer_bulk.RmdIntroduction
This vignette describes the workflow for running K2Taxonomer recursive partitioning on bulk gene expression data (Reed and Monti 2021). Note, that many of these steps are shared with that of single-cell expression analyses. A vignette for running K2Taxonomer on single-cell expression data can be found here.
Requirements
Read in sample ExpressionSet object
The main input of K2Taxonomer is an expression matrix object with approximately normally distributed expression values. Here we read in an example data set, which includes an expression matrix and sample data. See ?sample.ExpressionSet for more information about these data.
data(sample.ExpressionSet)Running K2Taxonomer
Get necessary data objects
Initialize K2 object
The K2preproc() initializes the K2 object
and runs pre-processing steps. Here, you can specify all arguments used
throughout the analysis. Otherwise, you can specify these arguments
within the specific functions for which they are implemented. See help
pages for more information.
A description of arguments implemented in this vignette are
- object: “Expression Matrix”: Expression matrix to be used for partioning and downstram analyses.
- colData: “Data Frame”: A data frame which contains a row for each profile (i.e. columns in object).
- genesets: “Named List”: A named list of gene sets to be used for downstream annotation.
Note, that many of the default arguments are chosen for the single-cell workflow. However, these will be replaced if the argument, cohort is not specified, and a message is printed.
## Run pre-processing
K2res <- K2preproc(expression_matrix,
colData = sample_data,
genesets = genesetsExample)
#> No cohorts specified and clustFunc = 'cKmeansDownsampleSqrt' . Setting clustFunc = 'hclustWrapper'
#> No cohorts specified and recalcDataMatrix = TRUE. Setting recalcDataMatrix = FALSE
#> No cohorts specified and featMetric = 'F'. Setting featMetric = 'mad'Perform recursive partitioning
The K2Taxonomer is
run by K2tax(). At each recursion of the algorithm, the
observations are partitioned into two sub-groups based on a compilation
of repeated K=2 clustering on bootstrapped sampling of features. For
each partition in the recursion, a stability metric is used to estimate
robustness, which takes on values between 0 and 1, where values close to
1 represent the instance in which the same clustering occured in every
or nearly every perturbation of a large set of observations. As the
number of observations decreasing down the taxonomy the largest possible
stability estimate decreases, such that the largest possible stability
estimate of triplets and duplets, is 0.67 and 0.50, respectively.
The parameter, stabThresh, controls the minimum
value of the stability metric to continue partitioning the observations.
By default, stabThresh is set to 0, which will run the
algorithm until until all observations fall into singlets. If we set
stabThresh=0.5 the algorithm can not separate duplets,
as well as larger sets that demonstrate poor stability when a partition
is attempted. This can also be set during initialization with
K2preproc().
Choosing an appropriate threshold is dependent on the size of the original data set. For large data sets, choosing small values will greatly increase runtime, and values between 0.8 and 0.7, are generally recommended.
## Run K2Taxonomer aglorithm
K2res <- K2tax(K2res,
stabThresh=0.5)Generate dendrogram from K2Taxonomer results
Static dendrogram
## Get dendrogram from K2Taxonomer
dendro <- K2dendro(K2res)
## Plot dendrogram
ggdendrogram(dendro)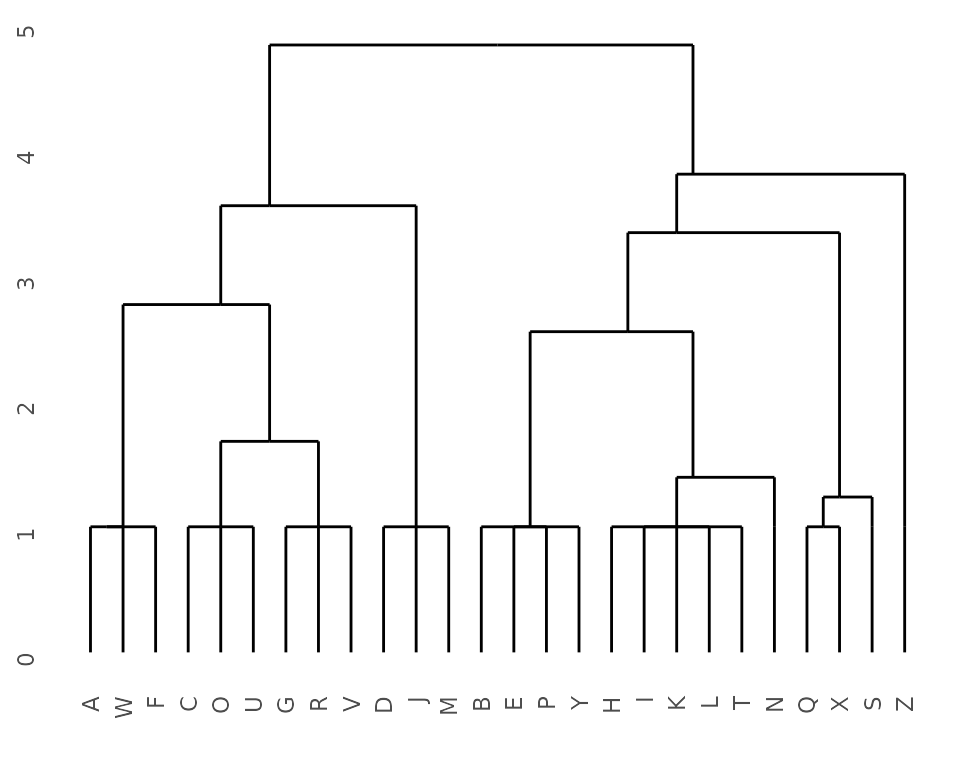
Interactive dendrogram
K2visNetwork(K2res)Annotate K2Taxonomer results
Partition-level Differential Gene Expression Analysis
K2res <- runDGEmods(K2res)
#> Running DGE for partition:
#> 1 / 10
#> 2 / 10
#> 3 / 10
#> 4 / 10
#> 5 / 10
#> 6 / 10
#> 7 / 10
#> 8 / 10
#> 9 / 10
#> 10 / 10Perform gene set based analyses
### Perform Fisher Exact Test based over-representation analysis
K2res <- runFISHERmods(K2res)
### Perform single-sample gene set scoring
K2res <- runScoreGeneSets(K2res)
#> Scoring gene sets with GSVA.
#> Estimating GSVA scores for 3 gene sets.
#> Estimating ECDFs with Gaussian kernels
### Perform partition-evels differential gene set score analysis
K2res <- runDSSEmods(K2res)
#> Running different enrichment score for partition:
#> 1 / 10
#> 2 / 10
#> 3 / 10
#> 4 / 10
#> 5 / 10
#> 6 / 10
#> 7 / 10
#> 8 / 10
#> 9 / 10
#> 10 / 10Explore Annotation Results
Partition-level Differential Gene Expression Analysis
Create Static Table of DGE Results
DGEtable <- getDGETable(K2res)
head(DGEtable)
#> gene coef mean t df pval fdr edge
#> 1 31583_at 1444.0457 2819.3481 7.090895 27.31219 1.184936e-07 0.0003465938 2
#> 2 31546_at 1180.8349 1963.0257 7.019826 27.31219 1.419856e-07 0.0004151659 2
#> 3 31527_at 1579.2611 3428.9188 6.704309 27.31219 3.191638e-07 0.0009329158 2
#> 4 31568_at 1372.3639 2866.9177 6.494425 27.31219 5.503484e-07 0.0016081181 2
#> 5 31573_at 1155.9527 2034.8062 6.331862 27.31219 8.418968e-07 0.0024591805 2
#> 6 31492_at 267.2776 307.9889 6.314658 27.31219 8.807715e-07 0.0025718527 2
#> node direction
#> 1 A up
#> 2 A up
#> 3 A up
#> 4 A up
#> 5 A up
#> 6 A upCreate Interactive Table of DGE Results
getDGEInter(K2res, minDiff = 1, node = c("A"), pagelength = 10)Create Plot of Gene Expression
plotGenePathway(K2res, feature = "31583_at", node = "A", subsample = FALSE)
#> K2eMatDS() is empty, using K2eMat() insteadCreate Static Plot Gene Expression
plotGenePathway(K2res, feature = "31583_at", node = "A", use_plotly = FALSE)
#> K2eMatDS() is empty, using K2eMat() instead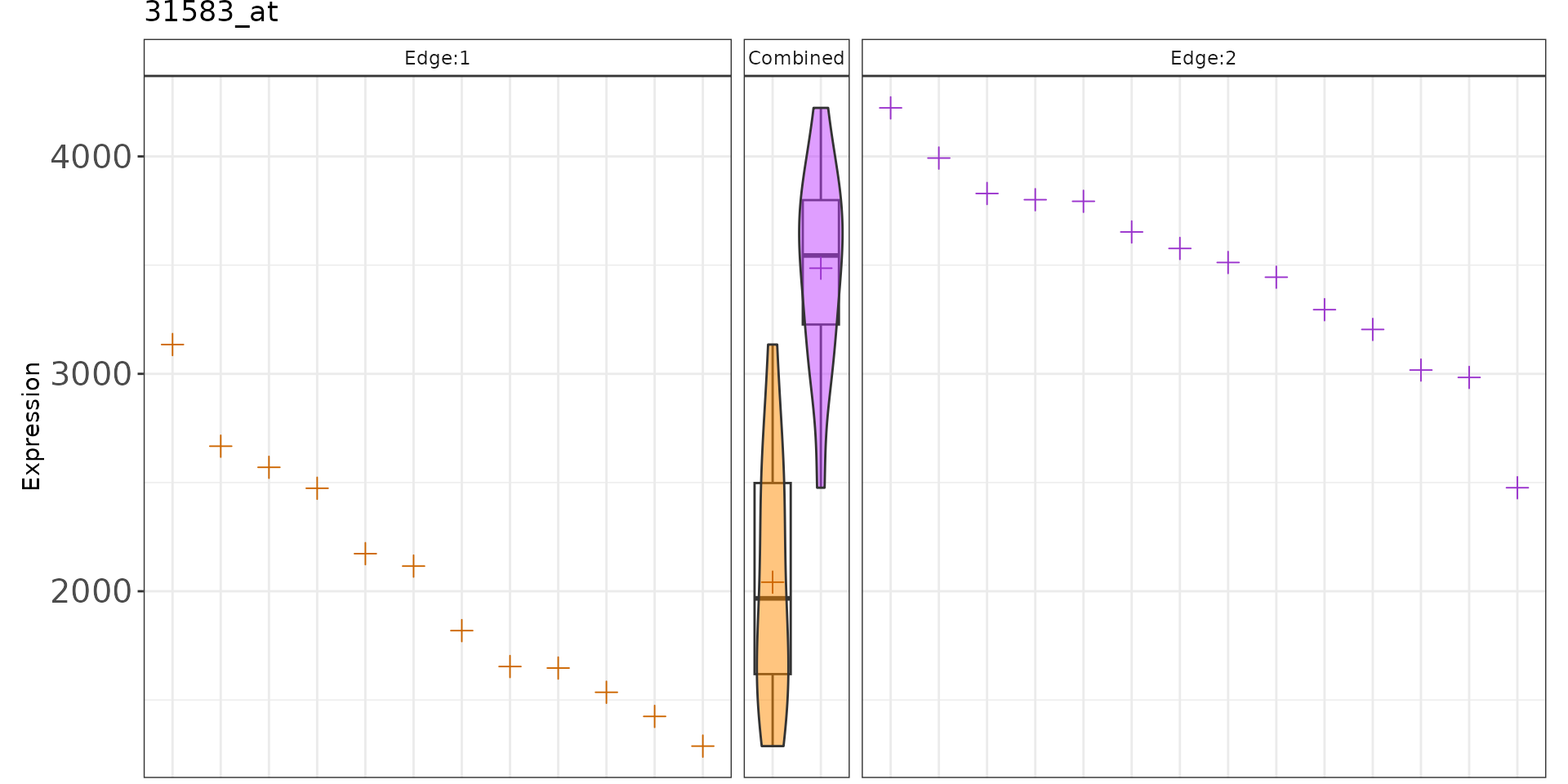
Partition-level Gene Set Enrichment Results
Create Static Table of Gene Set Results
ENRtable <- getEnrichmentTable(K2res)
head(ENRtable)
#> category node edge direction pval_fisher fdr_fisher nhits ndrawn ncats ntot
#> 1 GS1 G 2 up NA NA NA NA NA NA
#> 2 GS3 A 1 up NA NA NA NA NA NA
#> 3 GS1 A 1 up NA NA NA NA NA NA
#> 4 GS3 G 2 up NA NA NA NA NA NA
#> 5 GS1 B 1 up NA NA NA NA NA NA
#> 6 GS1 E 2 up 1 1 0 1 50 20000
#> pval_limma fdr_limma coef mean t hits
#> 1 0.004463359 0.08034045 0.3012940 0.171766848 3.954464 <NA>
#> 2 0.004788253 0.08140029 0.1851625 0.008112864 2.985053 <NA>
#> 3 0.009477649 0.15164239 0.1956683 -0.001944512 2.723717 <NA>
#> 4 0.081900911 1.00000000 0.2107563 0.112850770 1.997425 <NA>
#> 5 0.130129246 1.00000000 0.2138517 0.103415335 1.563280 <NA>
#> 6 0.179713417 1.00000000 0.1191463 -0.100852195 1.370722Create Interactive Table of Gene Set Results
getEnrichmentInter(K2res, nodes = c("A"), pagelength = 10)Create Interactive Plot of Single-sample Scoring
plotGenePathway(K2res, feature = "GS1", node = "A", type = "gMat")Create Static Plot of Single-sample Scoring
plotGenePathway(K2res, feature = "GS1", node = "A", type = "gMat", use_plotly = FALSE)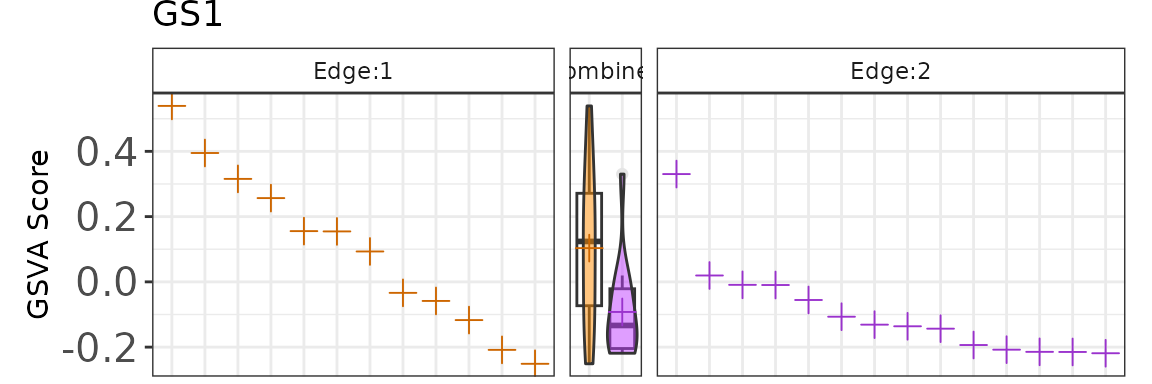
Create Dashboard
For more information about K2Taxonomer dashboards, read this vignette.
# Not run
K2dashboard(K2res, "K2results_sample.ExpressionSet")Implementation Options
Parellel excecutation
The K2Taxonomer workflow can take a long time with large data sets.
Accordingly, it is generally recommended to run the workflow using
parallel computing. This can be implemented easily by setting the
useCors argument in K2preproc()
# Not run
K2res <- K2preproc(expression_matrix,
colData = sample_data,
genesets = genesetsExample,
stabThresh=0.5,
useCors = 8) ## Runs K2Taxonomer in parellel with eight cores.Using ExpresssionSet object directly
In addition to expression matrices, ExpressionSet
objects may be input directly with the object argument.
When implemented, colData isn’t specified and this
information is pulled from the phenotype data of the
ExpressionSet object.
K2res_eSet <- K2preproc(sample.ExpressionSet,
genesets = genesetsExample,
stabThresh=0.5)
#> No cohorts specified and clustFunc = 'cKmeansDownsampleSqrt' . Setting clustFunc = 'hclustWrapper'
#> No cohorts specified and recalcDataMatrix = TRUE. Setting recalcDataMatrix = FALSE
#> No cohorts specified and featMetric = 'F'. Setting featMetric = 'mad'
## Run recursive partitioning algorithm
K2res_eSet <- K2tax(K2res_eSet)
## Get dendrogram from K2Taxonomer
dendro_eSet <- K2dendro(K2res_eSet)
## Plot dendrogram
ggdendrogram(dendro_eSet)