K2Taxonomer Dashboards
Eric R. Reed
March 25, 2025
Source:vignettes/03_K2Taxonomer_dashboard.Rmd
03_K2Taxonomer_dashboard.RmdIntroduction
To facilitate the comprehensive interrogation the output of K2Taxonomer this package includes functionality for generating interactive dashboards, which include full compendia of results (Reed and Monti 2021). In addition to the partition-level molecular comparisons included in these dashboards, they include functionality for performing molecular comparisons of gene expression and gene set enrichment on user-specified sets of two-or-more subgroups. In this vignette we describe the steps for creating, editing, and navigating interactive dashoboards from the output of the K2Taxonomer workflow (Reed and Monti 2021). For a more detailed description for running K2Taxonomer visit the vignettes describing single-cell and single-cell workflows here, and here, respectively.
Run K2Taxonomer workflow (Single-cell Example)
Read in single-cell RNAseq data
data("ifnb_small")Read in gene sets for subgroup annotation
data("cellMarker2_genesets")Get necessary data objects
## Integrated expression matrix used for clustering data
integrated_expression_matrix <- ifnb_small@assays$integrated$scale.data
## Normalized expression matrix to be used for downstream analyses
normalized_expression_matrix <- ifnb_small@assays$SCT$data
## Profile-level information
cell_data <- ifnb_small@meta.dataRun K2Taxonomer workflow
# Initialize `K2` object
K2res <- K2preproc(object = integrated_expression_matrix,
eMatDS = normalized_expression_matrix,
colData = cell_data,
cohorts="cell_type",
nBoots = 200,
clustFunc = "cKmeansDownsampleSqrt",
genesets = cellMarker2_genesets)
# Perform recursive partitioning
K2res <- K2tax(K2res)
# Partition-level differential gene expression
K2res <- runDGEmods(K2res)
### Perform Fisher Exact Test based over-representation analysis
K2res <- runFISHERmods(K2res)
### Perform single-sample gene set scoring
K2res <- runScoreGeneSets(K2res)
### Perform partition-level differential gene set score analysis
K2res <- runDSSEmods(K2res)Check partitioning results
## Get dendrogram from K2Taxonomer
dendro <- K2dendro(K2res)
## Plot dendrogram
ggdendrogram(dendro)Creating a dashboard
We generate the interactive dashboards with the
K2dashboard() function. This function will create a
directory with two files. These two files include the interactive
RMarkdown (.Rmd) file created by the flexdashboard
package and the R object (.rds) file containing the K2
object created by K2Taxonomer
functions.
Below we shown the K2dashboard() function and each of
its arguments.
The first argument, specifying the K2 object is the only
requirement. The remaining three arguments costumize the output, as
follows:
analysis_name: Specifies the title to be printed on the top of the dashboard. Also, specifies the prefix of the name of the output files. For filenames, spaces are replaced by underscores.
about: Logical specifying whether to include an about page in the dashboard. If TRUE (default), a third file is written to the dashboard directory, “about.md”. This is an editable markdown (.md) file, which the user is free to edit, and includes some default information about where to learn more about K2Taxonomer and how to navigate the dashboard. Customizing this file is described in more detail below.
output_dir: This is simply the path to the directory to write the dashboard directory.
K2dashboard(K2res,
analysis_name="K2Taxonomer Example",
about=TRUE,
output_dir=".")Additional considerations
The output directory
To prevent the overwriting of dashboard files, in addition to the analysis_name, the name of dashboard directory includes the date and time. It is formatted as follows:
Note: Changing the name of this directory does not effect the dashboard.
The “about.md” file
K2Taxonomer allows users to include information about their study in the interactive dashboards by editing” the “about.md” markdown file. When the “about.md” is included in the dashboard directory, it is read in and the markdown code is compiled along with the code in the dashboard file.
This file uses markdown syntax specific to the CRANpkg(“flexdashboard”) “Multiple Pages” layout, which can be reviewed here. Mainly, the first two lines of the default “about.md” file.
about
=====================================Will result in a tab, named About, as the first tab in the dashboard. Furthermore, lines with headers should start at three hashes, “###”.
Finally, when including links in the “about.md” file, make sure that clicking on these links opens a new window using the following:
[TEXT](URL){target="_blank"}Otherwise, the dashboard will need to reload upon navigating back.
Navigating dashboards
K2Taxonomer dashboards comprise three tabs, described below:
About: This optional tab with which the user can include information about the analysis being performed. This page is populated by a file, “about.md”, included in the same directory as the dashboard file. More information about formatting this file can be found here.
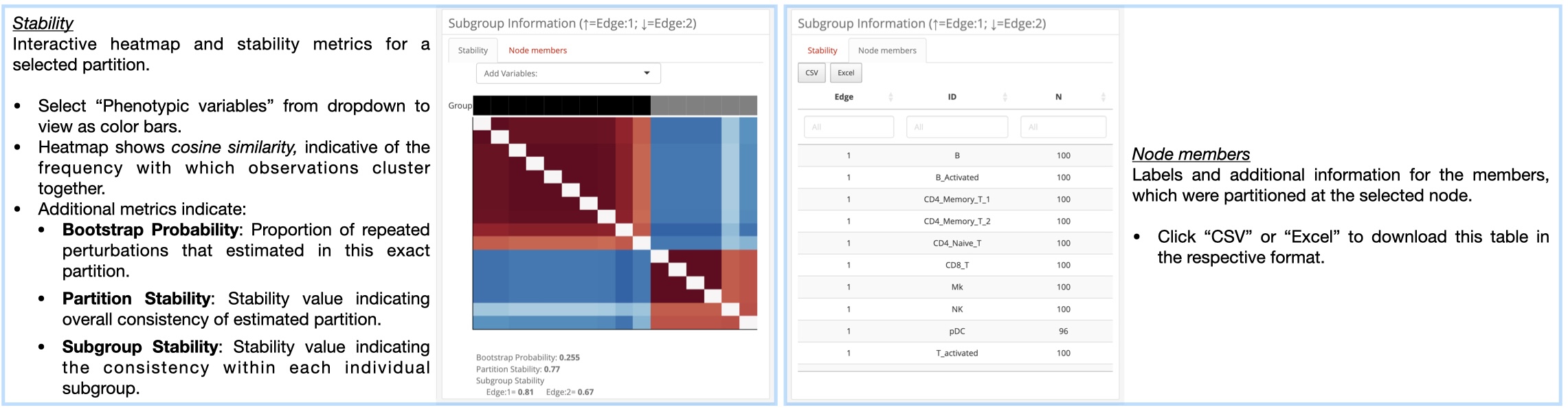
K2Taxonomer Results: This tab includes all of the results generated throughout the K2Taxonomer workflow, including: partitioning results, partition stability, gene expression analysis, gene set enrichment, and phenotypic variable testing (optional). More information for how K2Taxonomer estimates these results can be found here.
Compare Multiple: This tab allows the user to perform additional molecular comparisons between subgroups, beyond the partition-level comparisons performed by K2Taxonomer functions.
Annotation of each of these tabs is presented below. Click on any of these images to view in higher resolution