This is a simple example, where we generate data from a given linear model (with known intercept and slope), and then we apply linear regression to estimate the parameters of the data generating model.
set.seed(159) # for reproducible results
nobs <- 1000 # sample size
beta0 <- 1 # true intercept
beta1 <- 0.15 # true slope
## simulate an imaginary dependent variable (e.g., age between 15-75)
X <- sample(15:75,nobs,replace=TRUE)
Y <- rnorm(nobs,mean=beta0 + beta1 * X,sd=1)
## or, equivalently
## Y <- beta0 + beta1 * X + rnorm(nobs,mean=0,sd=1)
## png(file.path(OMPATH,"Rmodules/figures/diffanalLM.png"))
par(mar=c(c(5, 4, 4, 5) + 0.1))
plot(X,Y,pch=20,xlab="age",ylab="expression")
abline(beta0,beta1,col="red",lty=3,lwd=2)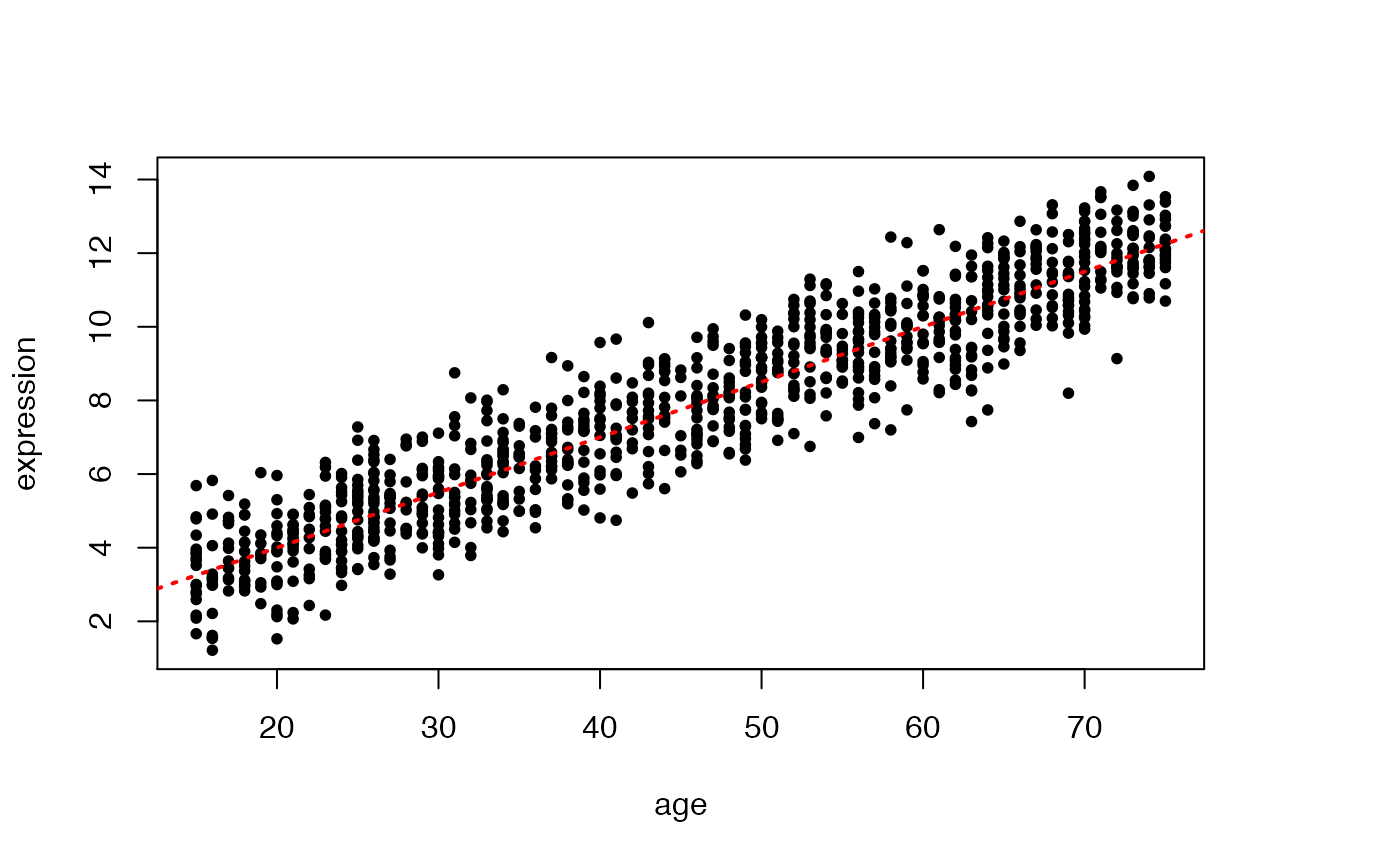
## notice the use of 'expression' to display mathematical symbols
##text(50,2,labels=expression(Y=beta[0]+beta[1]*X),las=1)We now fit a linear model to the generated data by lm.
##
## Call:
## lm(formula = Y ~ X)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.2064 -0.6992 -0.0125 0.6870 3.0741
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.004106 0.088484 11.35 <2e-16 ***
## X 0.150705 0.001812 83.18 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.014 on 998 degrees of freedom
## Multiple R-squared: 0.8739, Adjusted R-squared: 0.8738
## F-statistic: 6919 on 1 and 998 DF, p-value: < 2.2e-16As you can see, the estimates are quite close to the generating parameters.