The ExpressionSet Data Object
Stefano Monti
Source:vignettes/docs/ExpressionSet.Rmd
ExpressionSet.RmdSimple example of use of the R object ExpressionSet,
ideal for the storage of gene expression or similarly structured omic
data.
The ExpressionSet object
An expression set is a data object consisting of three entities: the
expression matrix (exprs), the phenotye data
(pData), and the feature data (fData).
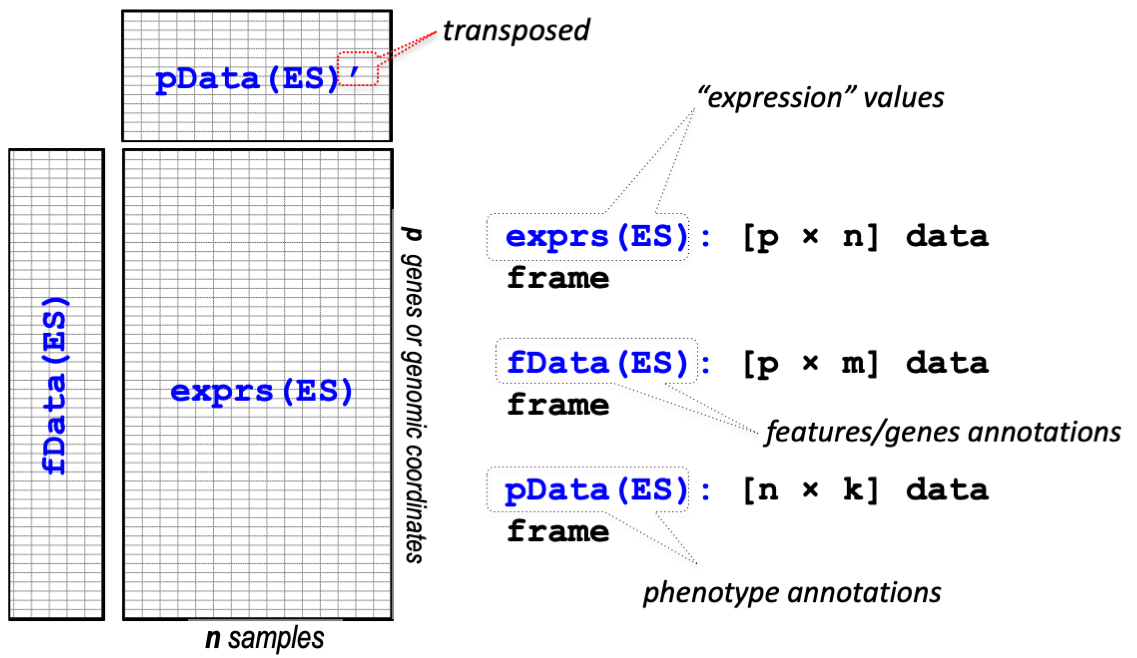 Figure 1: ExpressionSet object
Figure 1: ExpressionSet object
We upload an expression set already available. It corresponds to a subset of samples from a gene expression dataset of head and neck squamous carcinoma (HNSC) primary tissue samples from the TCGA project.
# Load toy dataset
data(HNSC_RNASeq_toy_ES)
## if BS831 not installed you can read the data from the Google Drive folder:
## HNSC_RNASeq_toy_ES <- readRDS(file.path(Sys.getenv("OMPATH"),"data/HNSC_RNASeq_toy_ES.rds"))
## rename for easier handling
hnsc <- HNSC_RNASeq_toy_ES
dim(hnsc) # the expression data## Features Samples
## 19844 25## [1] 25 6## bcr_patient_barcode bcr_sample_barcode tissue_type my_grade
## TCGA-CV-6962-11 TCGA-CV-6962 TCGA-CV-6962-11 AN AN
## TCGA-CV-7432-11 TCGA-CV-7432 TCGA-CV-7432-11 AN AN
## TCGA-CV-7177-11 TCGA-CV-7177 TCGA-CV-7177-11 AN AN
## TCGA-CV-7438-11 TCGA-CV-7438 TCGA-CV-7438-11 AN AN
## TCGA-CV-7440-11 TCGA-CV-7440 TCGA-CV-7440-11 AN AN
## TCGA-BA-7269-01 TCGA-BA-7269 TCGA-BA-7269-01 Tumor g1
## my_stage patient.anatomic_neoplasm_subdivision
## TCGA-CV-6962-11 AN larynx
## TCGA-CV-7432-11 AN oral cavity
## TCGA-CV-7177-11 AN larynx
## TCGA-CV-7438-11 AN oral tongue
## TCGA-CV-7440-11 AN larynx
## TCGA-BA-7269-01 stage iii oral tongue## [1] 19844 3## entrez_id gene_symbol
## ?|100130426 100130426 <NA>
## ?|100133144 100133144 <NA>
## ?|100134869 100134869 UBE2Q2P2
## ?|10357 10357 HMGB1P1
## ?|10431 10431 <NA>
## ?|155060 155060 LOC155060
## gene_description
## ?|100130426 <NA>
## ?|100133144 <NA>
## ?|100134869 ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 2
## ?|10357 high mobility group box 1 pseudogene 1
## ?|10431 <NA>
## ?|155060 AI894139 pseudogeneOne of the advantages of using an ExpressionSet is that the three component objects are always properly paired, and subsetting can be carried out straightforwardly.
tmp <- hnsc[1:100,1:10]
dim(tmp) # the expression data## Features Samples
## 100 10## [1] 10 6## [1] 100 3The SummarizedExperiment object
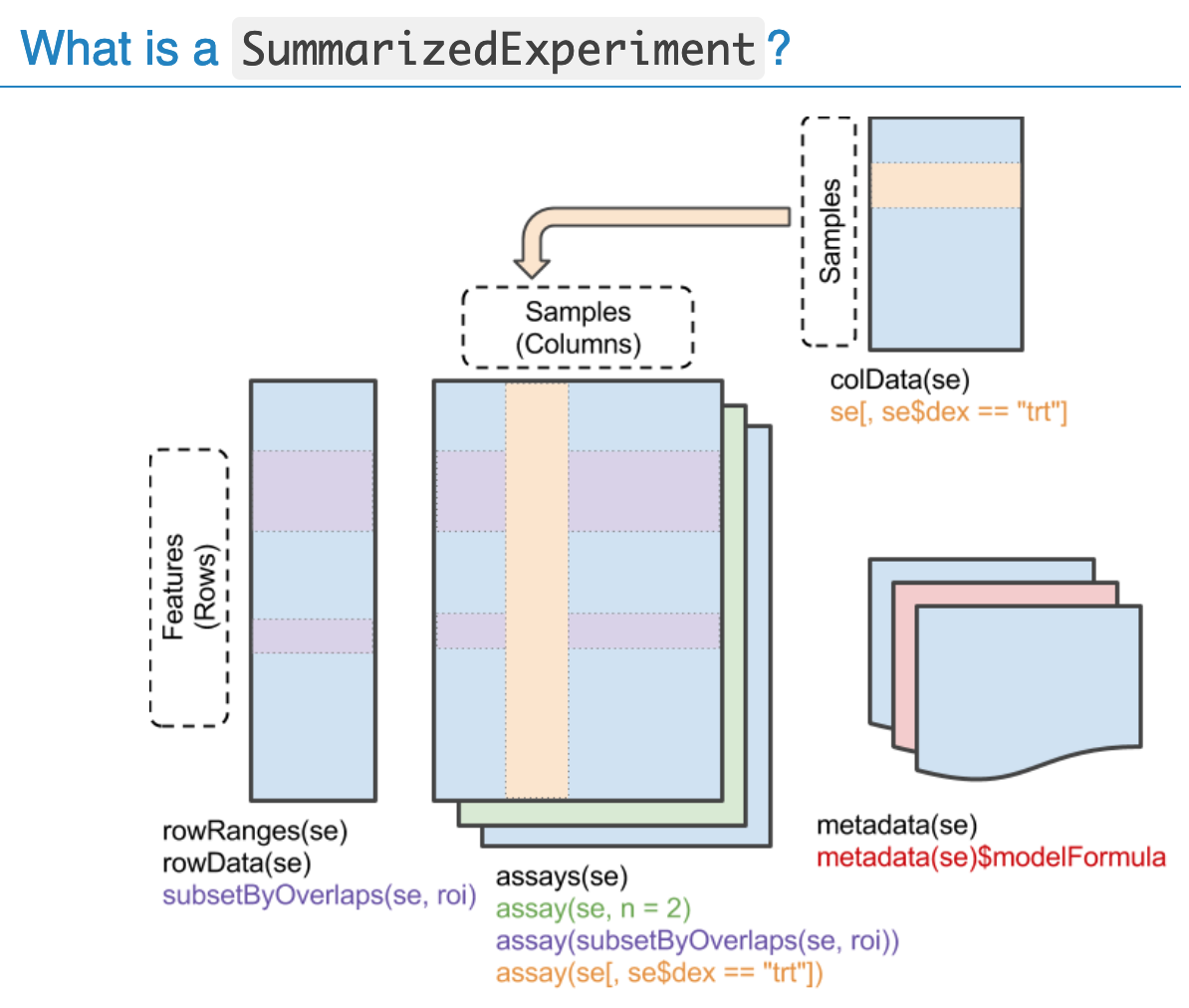 Figure 2:
SummarizedExperiment object [source].
Figure 2:
SummarizedExperiment object [source].
The ExpressionSet is generally used for array-based
experiments and gene expression data, where the rows are features, and
the SummarizedExperiment is generally used for
sequencing-based experiments, where the rows are GenomicRanges.
Mapping from ExpressionSet to
SummarizedExperiment
It is possible to easily map an ExpressionSet to a
SummarizedExperiment.
library(SummarizedExperiment)
sexp <- SummarizedExperiment::makeSummarizedExperimentFromExpressionSet(hnsc)
names(assays(sexp))## [1] "exprs"## DataFrame with 6 rows and 6 columns
## bcr_patient_barcode bcr_sample_barcode tissue_type my_grade
## <factor> <factor> <character> <character>
## TCGA-CV-6962-11 TCGA-CV-6962 TCGA-CV-6962-11 AN AN
## TCGA-CV-7432-11 TCGA-CV-7432 TCGA-CV-7432-11 AN AN
## TCGA-CV-7177-11 TCGA-CV-7177 TCGA-CV-7177-11 AN AN
## TCGA-CV-7438-11 TCGA-CV-7438 TCGA-CV-7438-11 AN AN
## TCGA-CV-7440-11 TCGA-CV-7440 TCGA-CV-7440-11 AN AN
## TCGA-BA-7269-01 TCGA-BA-7269 TCGA-BA-7269-01 Tumor g1
## my_stage patient.anatomic_neoplasm_subdivision
## <character> <factor>
## TCGA-CV-6962-11 AN larynx
## TCGA-CV-7432-11 AN oral cavity
## TCGA-CV-7177-11 AN larynx
## TCGA-CV-7438-11 AN oral tongue
## TCGA-CV-7440-11 AN larynx
## TCGA-BA-7269-01 stage iii oral tongue## DataFrame with 6 rows and 3 columns
## entrez_id gene_symbol
## <factor> <factor>
## ?|100130426 100130426 NA
## ?|100133144 100133144 NA
## ?|100134869 100134869 UBE2Q2P2
## ?|10357 10357 HMGB1P1
## ?|10431 10431 NA
## ?|155060 155060 LOC155060
## gene_description
## <factor>
## ?|100130426 NA
## ?|100133144 NA
## ?|100134869 ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 2
## ?|10357 high mobility group box 1 pseudogene 1
## ?|10431 NA
## ?|155060 AI894139 pseudogene