An extension of hypeR for network enrichment through random walks.
Data
data(ig)This package works with undirected igraph objects or extensions of igraph objects.
is(ig)The following vertex attributes are expected at a minimum.
-
name- Unique node labels -
symbol- Node symbols corresponding to the genesets you use
-
community- Pre-detected community labels
-
color- Community-specific colors
head(igraph::as_data_frame(ig, what="vertices")) community color symbol name
GMPS 8 #3E134F GMPS GMPS
PDLIM1 15 #9A9800 PDLIM1 PDLIM1
JUNB 15 #9A9800 JUNB JUNB
RER1 17 #50E2BB RER1 RER1
ICAM1 14 #A96C00 ICAM1 ICAM1
NPFFR2 14 #A96C00 NPFFR2 NPFFR2
set.seed(1)
layout <- igraph::layout_with_graphopt(ig,
start=NULL,
niter=1000,
charge=0.005,
mass=30,
spring.length=0,
spring.constant=1,
max.sa.movement=5)
par(mar=c(0,0,0,0))
plot(ig,
vertex.size=4,
vertex.color="grey",
vertex.frame.color="black",
vertex.label=NA,
edge.width=1,
layout=layout)
Network characterization through random walks
library(hypeR)
genesets <- hypeR::msigdb_gsets("Homo sapiens", "H", clean=TRUE)
ig.e <- enrich_communities_static(ig, genesets, restart=0.5, fdr=0.01, top=1)
par(mar=c(0,0,0,0))
set.seed(1234)
plot(ig.e,
vertex.size=5,
vertex.label=V(ig.e)$enrichment,
vertex.color=adjustcolor(V(ig.e)$color, alpha.f=0.6),
vertex.frame.color=adjustcolor("#000000", alpha.f=0),
vertex.label.family="Helvetica",
vertex.label.color="black",
vertex.label.font=2,
vertex.label.cex=0.8,
vertex.label.dist=runif(vcount(ig.e), -0.25, 0.25),
edge.color=adjustcolor("#000000", alpha.f=0.2),
layout=layout)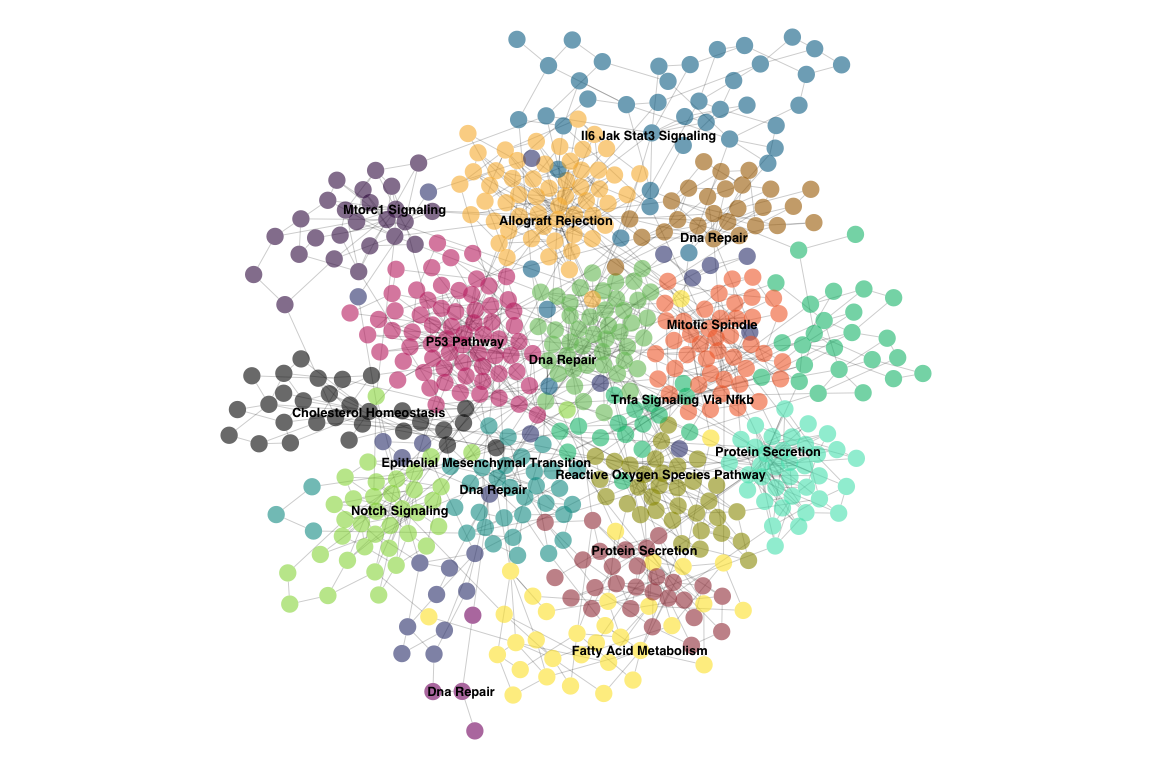
Please read the documentation for a comprehensive overview of functionality.

