Markov Networks-based Differential Connectivity Analysis
Ziwei Huang
10/06/2025
Source:vignettes/differential_connectivity_analysis.Rmd
differential_connectivity_analysis.RmdIntroduction
Differential connectivity analysis is an important application in network-based studies, aimed at characterizing network rewiring patterns of interacting entities across different conditions or phenotypes. Unlike univariate analyses that evaluate features independently, this approach captures integrative, system-level signals arising from the collective behavior of interacting entities, thereby providing complementary insights into underlying mechanisms.
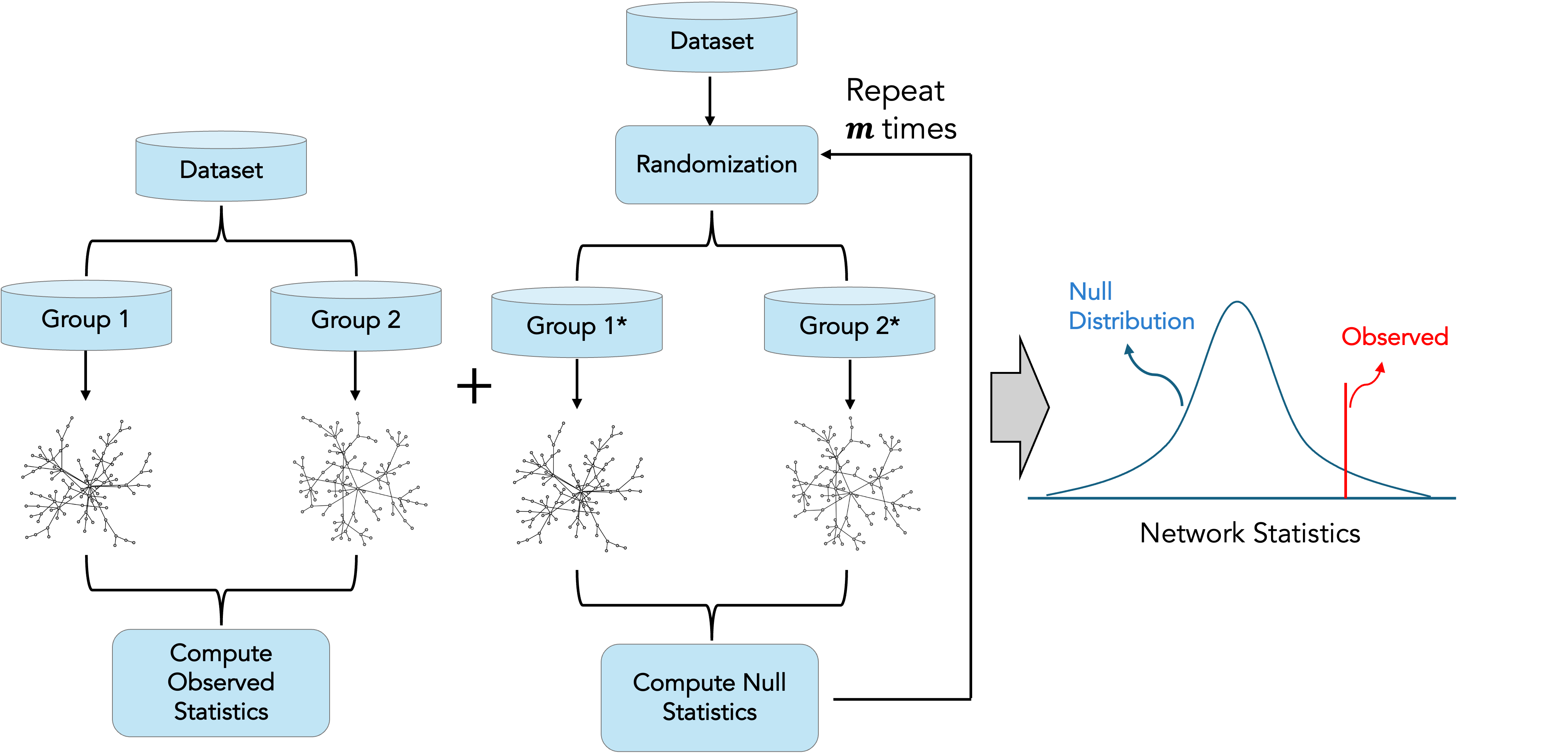
RSNet supports differential connectivity analysis for datasets containing samples from two groups (e.g., young vs. old, disease vs. healthy) and enables statistical assessment of a wide range of network-based statistics, including centrality measures and graphlet degree vector (GDV)–based distances.The overall workflow is illustrated in the following figure. Briefly, the input dataset is partitioned into two mutually exclusive subsets corresponding to the groups of interest. Network structure inference is then performed separately for each subset, and the observed differences in network-based statistics are computed.
To assess statistical significance, RSNet constructs a resampling-based empirical null distribution by randomizing the original dataset using permutation and/or bootstrap procedures. The significance of the observed network differences is subsequently evaluated by comparing the observed statistics to the corresponding null distributions.
Load LOAD dataset
Here, we use a curated dataset derived from a late-onset Alzheimer’s disease (LOAD) study.
Here we used a real, and curated dataset derived from late-onset Alzheimer’s disease (LOAD) study. The dataset includes 129 LOAD subjects and 101 control subjects. For demonstration and computational efficiency, we use only a subset of genes with the highest median absolute deviation (MAD).
data(toy_load)Estimate observed networks
The observed networks are constructed by splitting the dataset into two groups based on the phenotype of interest, LOAD versus Control. Each subset of the data is then used as input to the resampling-based framework implemented in RSNet. The resulting networks derived from each group are stored as a named list object for downstream analyses.
## split the data into two groups
ctrl_dat <- toy_load %>%
dplyr::filter(phenotype == "ctrl") %>%
dplyr::select(-phenotype)
load_dat <- toy_load %>%
dplyr::filter(phenotype == "load") %>%
dplyr::select(-phenotype)
## Infer observed networks from each group
ctrl_obs <- capture_all(ensemble_ggm(dat = ctrl_dat,
num_iteration = 1,
sub_ratio = 1,
method = "D-S_NW_SL",
n_cores = 1)) %>%
consensus_net_ggm(., filter = "pval", threshold = 0.05) %>%
{ .[["consensus_network"]] }
load_obs <- capture_all(ensemble_ggm(dat = load_dat,
num_iteration = 1,
sub_ratio = 1,
method = "D-S_NW_SL",
n_cores = 1)) %>%
consensus_net_ggm(., filter = "pval", threshold = 0.05) %>%
{ .[["consensus_network"]] }
## Create a list of observed networks
obs_networks <- list(ctrl = ctrl_obs,
load = load_obs)Estimate the null distribution
Accurate estimation of the null distribution is essential for
assessing statistical significance. owever, for most network-based
statistics, the null distribution is difficult to derive analytically.
RSNet addresses this challenge by implementing
resampling-based null distribution generation via
permutation and bootstrap methods.
These procedures are implemented in the null_ggm()
function.
Because null generation can be computationally intensive,
null_ggm() also supports parallel
computing for improved efficiency.
In the following example, we generate two null distributions, one using permutation and one using bootstrap, each with five iterations.
shuffle_iter <- 5
null_permutation <- capture_all(null_ggm(dat = toy_load, # a n x (p+1) data frame with p numeric feature columns + one label column.
group_col = "phenotype", # name of the label (phenotype) column
inference_method = "D-S_NW_SL", # the inference method
shuffle_method = "permutation", # shuffling method
shuffle_iter = shuffle_iter, # number of iteration
filter = "pval", # filter criteria, support "pval", "fdr", and "none"
threshold = 0.05, # the significance level
n_cores = 1)) # number of cores for parallel computing
null_bootstrap <- capture_all(null_ggm(dat = toy_load,
group_col = "phenotype",
inference_method = "D-S_NW_SL",
shuffle_method = "bootstrap",
shuffle_iter = shuffle_iter,
filter = "pval",
threshold = 0.05,
n_cores = 1))
## A list of length "shuffle_iter"; each element is a named list (by group labels) of igraph objects
stopifnot(length(null_permutation$null_networks) == shuffle_iter)
stopifnot(length(null_bootstrap$null_networks) == shuffle_iter)Differential centralities
RSNet supports differential analysis of standard
centrality measures, including degree, eigenvector, and betweenness
centralities, etc. This functionality is implemented in the
diff_centrality() function, which takes as input the
observed networks。
Internally, diff_centrality() calls the
null_ggm() function to generate the null distribution. For
improved memory efficiency, users can alternatively run
null_ggm() separately and supply its output to the
null_networks argument.
The diff_centrality() function supports both
permutation-based and bootstrap-based
randomization procedures, and allows for one-sided or
two-sided hypothesis testing.
diff_centrality_res <- capture_all(diff_centrality(obs_networks = obs_networks,
dat = toy_load,
group_col = "phenotype",
null_networks = NULL,
alternative = "two.sided",
inference_method = "D-S_NW_SL",
shuffle_method = "permutation",
shuffle_iter = 5,
balanced = FALSE,
filter = "pval",
threshold = 0.05,
n_cores = 1,
seed = 42))
diff_centrality_res %>%
head(.) %>%
DT::datatable(.)Differential graphlet degree vector(GDV)-based distance
In addition to differential analysis of standard centrality measures,
RSNet also supports graphlet degree vector
(GDV)-based distance analysis (see the Graphlet page
for details). This functionality is implemented in the
diff_gdv() function.
The arguments of diff_gdv() are similar to those of
diff_centrality(). However, diff_gdv()
currently supports only permutation-based randomization
procedures and performs one-sided hypothesis testing,
consistent with the statistical characteristics of the GDV distance
measure.
diff_gdv_res <- capture_all(diff_gdv(obs_networks = obs_networks,
dat = toy_load,
group_col = "phenotype",
null_networks = NULL,
sign = TRUE,
inference_method = "D-S_NW_SL",
shuffle_method = "permutation",
shuffle_iter = 5,
balanced = FALSE,
filter = "pval",
threshold = 0.05,
n_cores = 1,
seed = 42))
diff_gdv_res %>%
head(.) %>%
DT::datatable(.)