Conditional Gaussian Bayesian Networks Structure Learning
Ziwei Huang
10/06/2025
Source:vignettes/cgbn.Rmd
cgbn.RmdIntroduction
A Conditional Gaussian Bayesian Network (CGBN) is a probabilistic graphical model that integrates discrete and continuous variables within a unified Bayesian framework, making it particularly suitable for mixed-type data. CGBNs are represented as directed acyclic graphs (DAGs), where nodes correspond to random variables and directed edges encode conditional dependencies consistent with a joint probability distribution in which continuous variables follow Gaussian distributions conditional on their discrete parents.
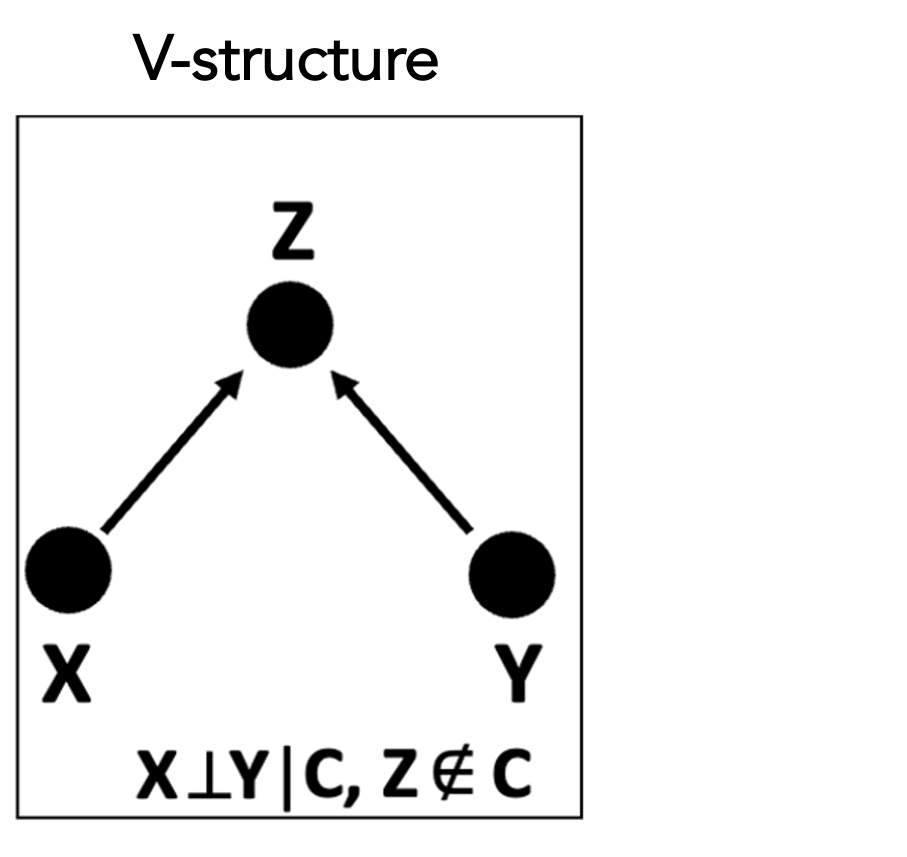
Structure learning in CGBNs is typically performed using constraint-based methods, which infer the network topology by testing conditional independence relationships implied by the data. Starting from a complete graph, edges are iteratively removed when conditional independence between variable pairs is detected given appropriate conditioning sets. A key step in orienting edges in the resulting partially directed graph is the identification of v-structures, of the form . A v-structure is inferred when two variables and are marginally independent but become conditionally dependent upon conditioning on a third variable , as illustrated in the following figure:
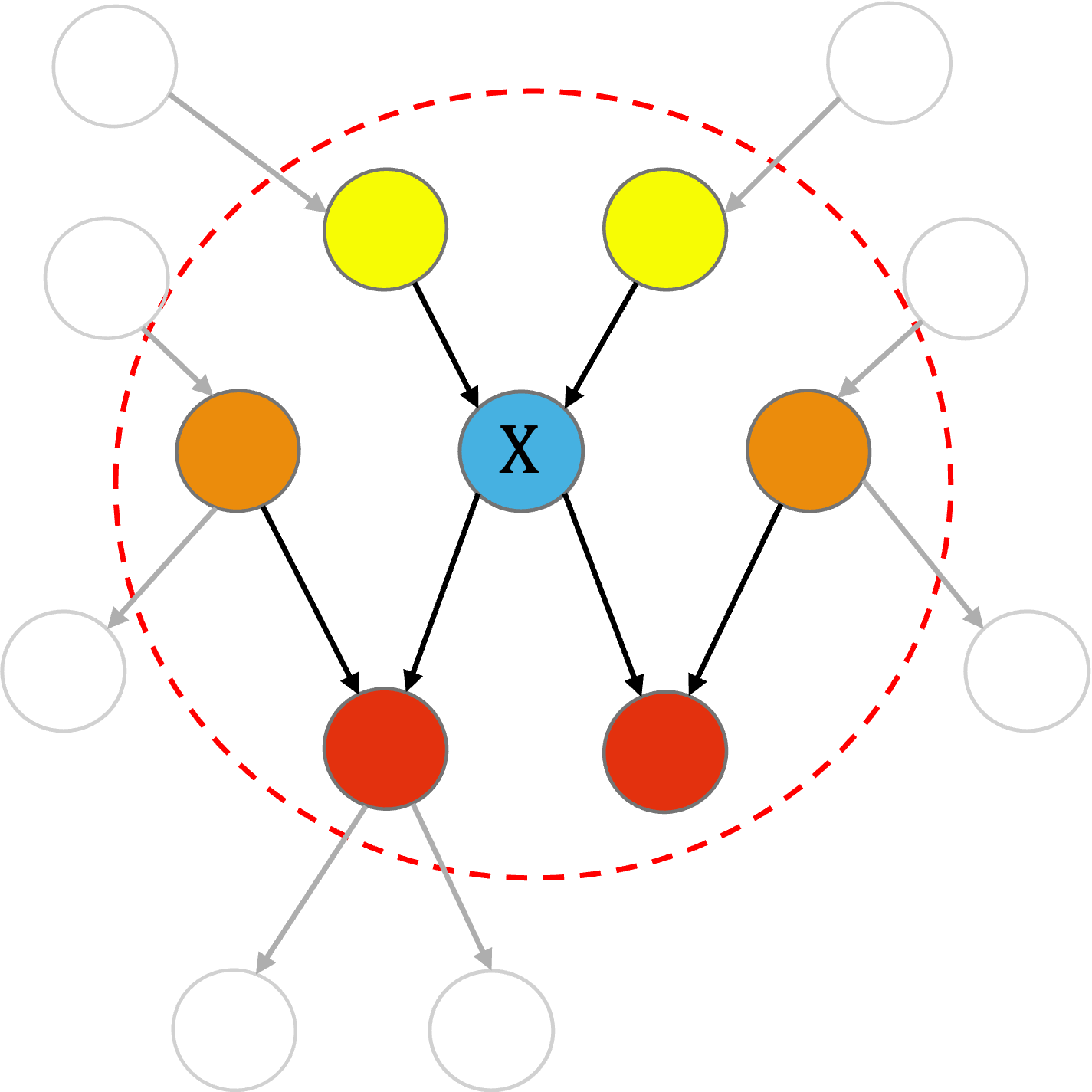
An important concept in CGBNs is the Markov blanket of a node, defined as the minimal set of variables that renders the node conditionally independent of all other variables in the network. For a given node, its Markov blanket consists of its parents, its children, and its spouses (i.e., the other parents of its children). As illustrated in the following figure, we use yellow, orange, and red to denote the parents, spouses, and children of node , respectively.
RSNet implements a resampling-based structure learning framework for CGBNs, supporting four resampling strategies to improve the stability and robustness of inferred network structures:
- Bootstrap.
- Sub-sampling
- Stratified bootstrap.
- Stratified sub-sampling.
IMPORTANT NOTE: The Conditional Gaussian Bayesian
Network functionalities ensemble_cgbn() and
consensus_net_cgbn() are optional in RSNet
and require the RHugin package. Installation instructions
for macOS,
windows,
and Linux.
Load a toy dataset
To illustrate the workflow, we use a synthetic toy dataset containing 20 continuous and 5 discrete variables, representing a mixed-type dataset suitable for conditional Gaussian Bayesian network analysis.
data("toy_cgbn")Run and learn an ensemble of networks from resampled datasets
In this example, we use the simulated dataset as input and perform
bootstrap resampling (boot = TRUE) with 5 iterations
(num_iteration = 5). The column names of the discrete
variables must be specified as a character vector in the
discrete_variable argument.
To perform subsampling instead of
bootstrapping, set boot = FALSE and
specify the sampling proportion using the sub_ratio
argument (a value between 0 and 1).
The function ensemble_cgbn() also supports parallel
computing.
ensemble_toy <- ensemble_cgbn(dat = toy_cgbn, # A n x p dataframe
discrete_variable = sprintf("D%d",1:5), # Column names of the discrete variables
num_iteration = 5, # Number of resampling iteration
sample_class = NULL, # Optional: for stratified sampling
boot = TRUE, # If FALSE, perform sub-sampling
sub_ratio = 1, # Subsampling ratio (0–1)
n_cores = 1) # Number of cores for parallel computingConsensus network construction
The construction of the consensus network for conditional Gaussian Bayesian networks (CGBNs) supports two complementary approaches:
method = "all": Computes the selection frequency of each edge (i.e., the proportion of resampling iterations in which the edge is identified) with respect to a reference network (typically inferred using all samples). The resulting consensus network retains its directed structure.method = "average": Computes the selection frequency of each edge based on Markov blanket relationships between the two incident nodes. The resulting consensus network is undirected, representing stable dependency patterns rather than directionality.
## Directed consensus network (method = "all")
## A simple way to generate a reference network is to set: num_iteration = 1, boot = FALSE, sub_ratio = 1
reference_network <- ensemble_cgbn(dat = toy_cgbn,
discrete_variable = sprintf("D%d",1:5),
num_iteration = 1,
sample_class = NULL,
boot = FALSE,
sub_ratio = 1,
n_cores = 1)
directed_cons_net <- consensus_net_cgbn(ensemble_toy$ig_networks, # A list of igraph objects
reference_network = reference_network$ig_networks$iter_1, # An igraph object
method="all", # Integration method
cut = 0.5) # Edge weight threshold
## Markov blanket-based undirected consensus network, set `method="average"
undirected_cons_net <- consensus_net_cgbn(ensemble_toy$ig_networks, # A list of igraph objects
reference_network = NULL, # Not required for method = "average"
method="average", # Integration method
cut = 0.5) # Edge weight threshold