A list of objects returned from candidate_search
using simulated dataset FS = sim_FS, input_score = sim_Scores,
top_N = 7, method = "ks_pval", alternative = "less",
search_method = "both", max_size = 10, and
best_score_only = FALSE.
data(topn_list)Format
A list of objects returned from candidate_search including
a set of best meta-feature matrix, its corresponding best score, its
observed input scores, rank of the best features based on their scores,
marginal best scores, and cumulative best scores. pertaining to each
top N feature searches.
See candidate_search for more information.
Value
a list of objects returned from candidate_search function
Details
NOTE: max_size is set to 10 as we would like to account
for the presence of 10 left-skewed (i.e. true positive or TP) features
in sim_FS dataset.
References
Kartha VK, Kern JG, Sebastiani P, Zhang L, Varelas X, Monti S (2019) CaDrA: A computational framework for performing candidate driver analyses using binary genomic features. (Frontiers in Genetics)
Examples
# Load pre-computed Top-N list generated for sim_FS and sim_Scores dataset
data(topn_list)
# Fetch the first meta-feature
topn_list[[1]]$feature_set
#> class: SummarizedExperiment
#> dim: 6 100
#> metadata(0):
#> assays(1): ''
#> rownames(6): TP_8 TP_9 ... TP_3 TP_7
#> rowData names(0):
#> colnames(100): 1 2 ... 99 100
#> colData names(0):
# Fetch the second meta-feature
topn_list[[2]]$feature_set
#> class: SummarizedExperiment
#> dim: 6 100
#> metadata(0):
#> assays(1): ''
#> rownames(6): TP_9 TP_8 ... TP_3 TP_7
#> rowData names(0):
#> colnames(100): 1 2 ... 99 100
#> colData names(0):
# Retrieve the meta-feature with the best score among top_N = 7 runs
topn_best_meta <- topn_best(topn_list = topn_list)
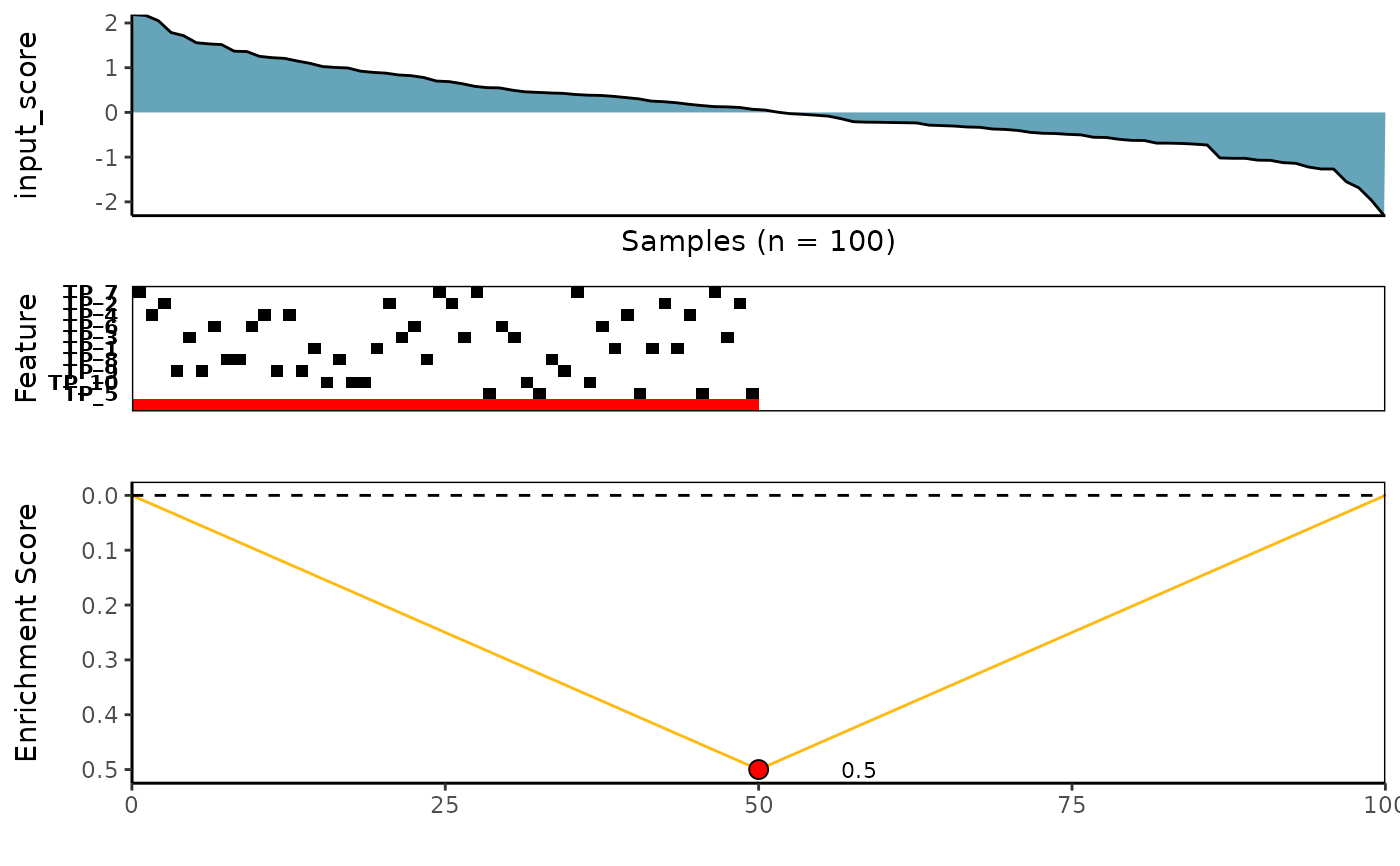
# Visualize the best meta-feature using meta_plot function
meta_plot(topn_best_list = topn_best_meta)
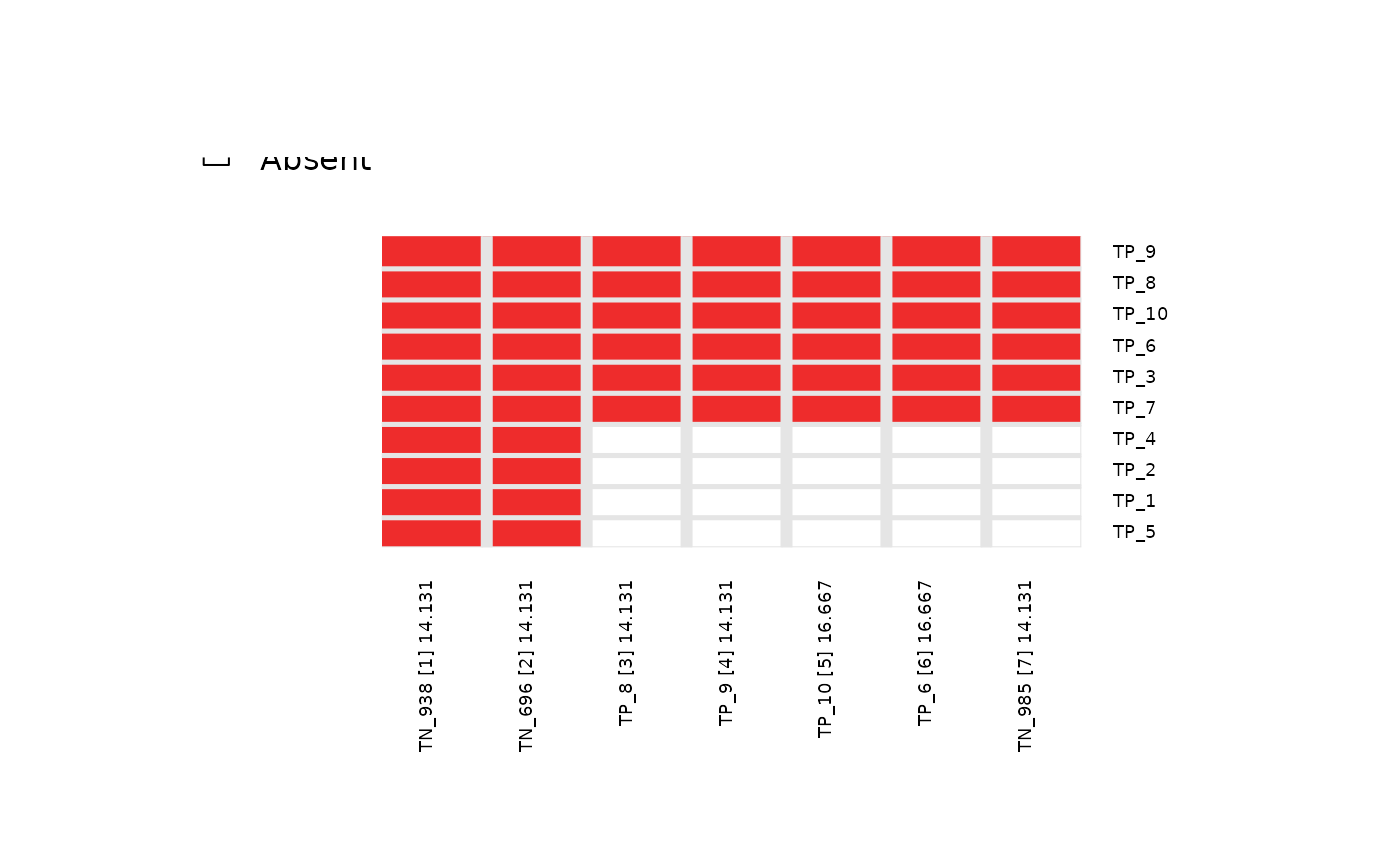
 # Visualize overlap of meta-features across top_N = 7
# using topn_plot function
topn_plot(topn_list = topn_list)
# Visualize overlap of meta-features across top_N = 7
# using topn_plot function
topn_plot(topn_list = topn_list)